# From the R console
base::R.version.string
# (This is the version on my own machine)
# [1] "R version 4.2.2 (2022-10-31)"Installation and setup
This workshop showcases introductory bio statistics concepts using the open source (and free!) programming language . Each session of the workshop features exercises that will help you learn by doing.
We will quickly go over this at our first Practice session, but you are expected to have the required software installed on your machine before the workshop.
Below is a step by step process that should guide you through the needed installation steps.
Install
R is available for free for Windows , GNU/Linux , and macOS .
- To install R, you can go to this link. The latest available release is R 4.4.1 “Race for Your Life” released on 2024/06/14, but any (fairly recent) version will do.
If you have previously installed R on your machine, you can check which version you are running by executing this command in R:
…or by executing this command in your CLI (Command Line Interface):
# From Terminal/Powershell/bash
R --versionInstall RStudio IDE
While not strictly required, it is highly recommended that you also install RStudio to facilitate your work. RStudio Desktop is an Integrated Development Editor (IDE), basically a graphical interface wrapping and interfacing R (which needs to be installed first).
R, which is a command line driven program, can be executed via its native interface (R GUI), as well as from many other code editors, like VS Code, Sublime Text, Jupyter Notebook, etc. RStudio remains the most widely used by beginners and advanced programmers alike, because of its intuitive and integrated interface.
- To install RStudio you can go to this link.
- The free RStudio Desktop version contains everything you need.
- Again, you don’t need to have the latest RStudio version, but I recommend v2022.07.1 or later (it also includes the installation of Quarto)
Quarto is a multi-language, next-generation version of R Markdown from Posit (former RSTUDIO company). While for R users it is not much different form Rmarkdown (you can author files with extension .qmd which are very similar), its key feature is that is It is available also for Python, Julia, and Observable languages.
Quarto is not an R package so it has to be downloaded separately. However Rstudio v2022.07.1 or later come with Quarto included
The www.r4statistics.com is actually a quarto website 😊
Install R packages from the CRAN
An R package is a shareable bundle of functions. Besides the basic built-in functions already contained in the R program (i.e. the base, stats, or utils packages), many more useful R functions come in free libraries of code (packages) written by the R users community.
- CRAN - the Comprehensive R Archive Network - is the general package repository for R: https://cran.r-project.org/
(We will only use packages from CRAN for our purposes)
Bioconductor - a package repository geared towards biostatistics: https://www.bioconductor.org/
Github - a repository where you can find developer’s version of a package: https://github.com/
R packages that will be required for the workshop
Below are the R packages, that we will need for the workshop’s practical sessions. You should have them pre-installed on your laptop ahead of the workshop as well (at least the ones needed fo Lab # 1: pkg_list_lab_1)… This will also serve as a test to check whether the R version you have is compatible.
A couple of things to be aware of when installing R packages:
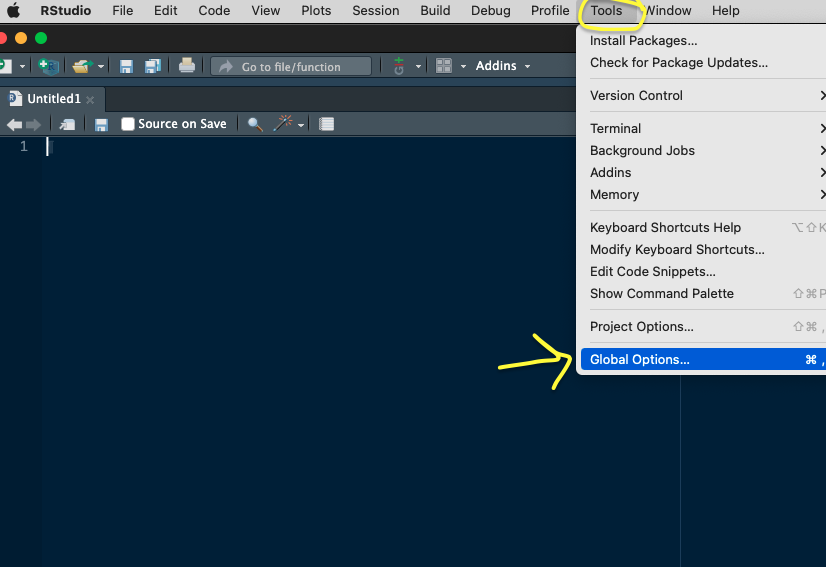
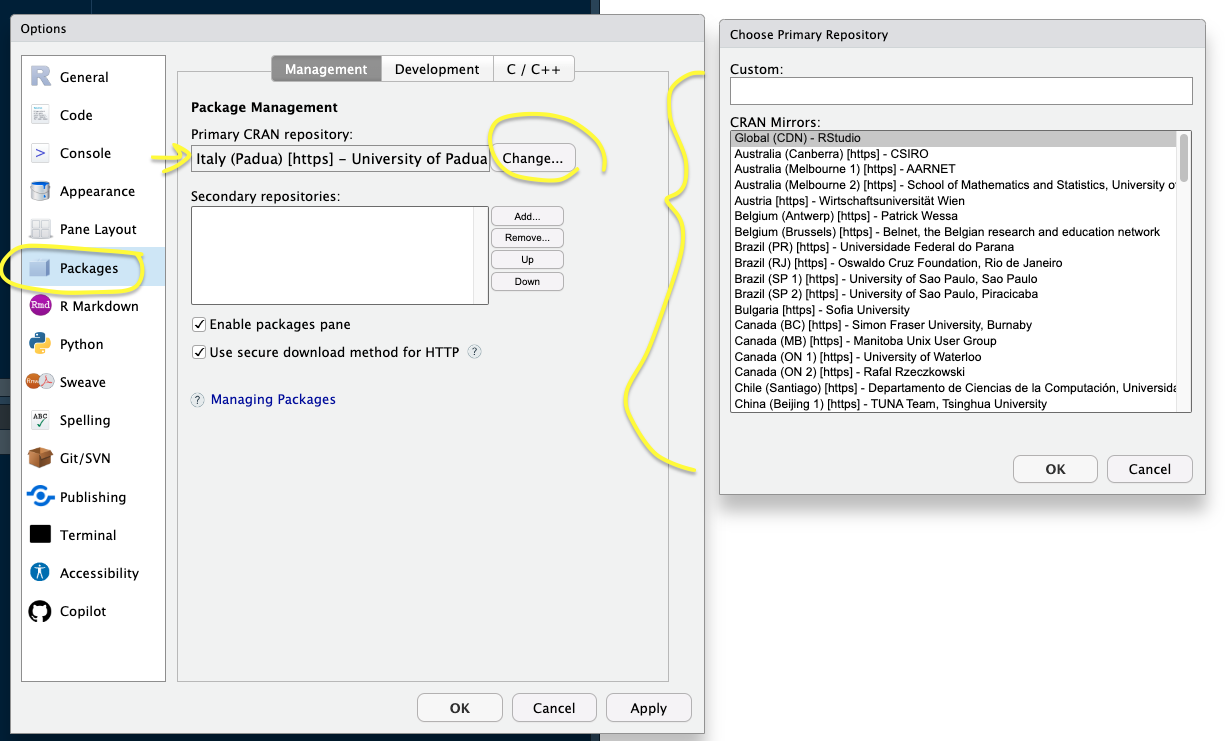
- By default, the RStudio IDE uses the RStudio CRAN global mirror (i.e. the primary repository with all the packages code globally distributed using Amazon S3 storage), but you could also override the default CRAN repository and choose another CRAN mirror (i.e. a server copy), perhaps one hosted at an institution near you – see box below.
R may automatically install some related packages if the package you are trying to install has what is known as a
dependency(other packages needed for it to work).-
R may ask you this question: “
Do you want to install from sources the package which needs compilation? (Yes/no/cancel). This (in most cases) means that the package has updated recently on CRAN but the binary isn’t yet available for your OS (can take a day or two).- To answer, follow instructions by responding directly in the R console (you are asked to type “Yes” or “No”):
- If you say “
no”, you won’t get the most recent version, and install from the pre-compiledbinariesavailable –which should be totally fine! (At least for our purposes). - If you say “
yes”, the package will be built from source locally. If itssource codeneeds compilation (i.e. has portions of code that need to be ‘translated’ from C/C++ or Fortran) and you’ve never set up build tools for R, then this may not succeed.
- If you say “
- To answer, follow instructions by responding directly in the R console (you are asked to type “Yes” or “No”):
At the end of the installation process you will get a message (bottom of the R Console) indicating were the source code for the package(s) has been stored on your machine.
Installing packages (1st time you use an R Package)
Option 1)
You could install one package at a time via the function install.packages()…
# (**ONLY** the 1st time you use them)
# Installing
install.packages("name_of_package_here" )
# [OPTIONAL ARGUMENT] Installing (with specification for dependencies)
install.packages("name_of_package_here" , dependencies = TRUE)… or in bulk, like so (it might take a few moments more):
# (**ONLY** the 1st time you use them)
# ---- Installing R pckgs for 1st LAB
pkg_list_lab_1 <- c("fs","here", "janitor", "skimr",
"dplyr", "forcats",
"ggplot2", "ggridges")
install.packages(pkg_list_lab_1)
# ---- Installing (more) R pckgs for 2nd LAB
pkg_list_lab_2 <- c("tidyr", "patchwork",
"ggthemes", "ggstatsplot", "ggpubr", "viridis",
"BSDA", "rstatix", "car", "multcomp")
install.packages(pkg_list_lab_2)
# ---- Installing (more) R pckgs for 3rd LAB
pkg_list_lab_3 <- c("openxlsx",
"lmtest" ,
"broom",
"performance")
install.packages(pkg_list_lab_3)
# ---- Installing (more) R pckgs for 4th LAB
pkg_list_lab_4 <- c("rsample",
"MASS",
"FactoMineR",
"factoextra",
"ggfortify",
"scatterplot3d",
"pwr" )
install.packages(pkg_list_lab_4)Option 2)
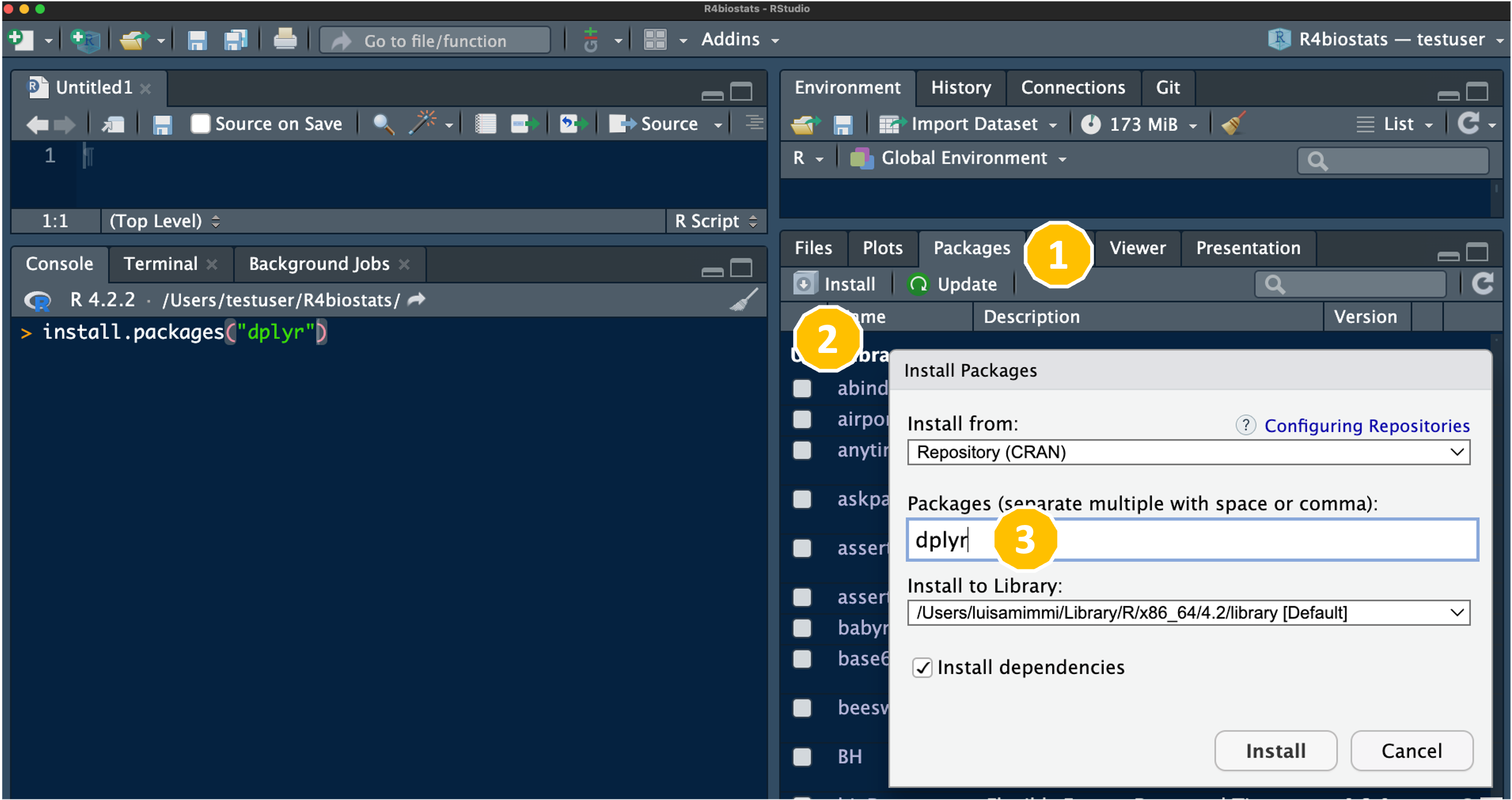
In alternative, you could install each package separately, using the RStudio GUI, from the Packages tab in the bottom right pane as indicated here:
Loading a package (at the beginning of every R session)
Once packages have been installed, with the command library() loads the specific R packages that you are going to need in any given R session.
# Loading a package (at the beginning of every R session)
# --- General
library(here) # tools find your project's files, based on working directory
library(fs) # file/directory interactions
library(janitor) # tools for examining and cleaning data
library(skimr) # Compact and Flexible Summaries of Data
library(openxlsx) # Read, Write and Edit xlsx Files
# --- Tidyverse
library(dplyr) # {tidyverse} A Grammar of Data Manipulation
library(tidyr) # {tidyverse} Tools to create tidy data
library(forcats) # {tidyverse} Tools for Categorical Var.(Factors)
# --- Plotting
library(ggplot2) # {tidyverse} tools for plotting
library(ggstatsplot)# 'ggplot2' Based Plots with Statistical Details
library(ggpubr) # 'ggplot2' Based Publication Ready Plots
library(patchwork) # Functions for ""Grid" Graphics"composing" plots
library(viridis) # Colorblind-Friendly Color Maps for R
library(ggthemes) # Extra Themes, Scales and Geoms for 'ggplot2'
library(ggridges) # Ridgeline Plots in 'ggplot2' (density functions)
# --- Statistics
library(BSDA) # Basic Statistics and Data Analysis
library(rstatix) # Pipe-Friendly Framework for Basic Statistical Tests
library(car) # Companion to Applied Regression
library(multcomp) # Simultaneous Inference in General Parametric Models
library(lmtest) # Testing Linear Regression Models
library(broom) # Convert Statistical Objects into Tidy Tibbles
library(performance)# Assessment of Regression Models Performance
library(pwr) # Basic Functions for Power AnalysisLearning about a package (after installation)
Once an R package is installed, you can also read the documentation about it directly inside the RStudio IDE. For example, try running in your Console:
# - To ask about a package
?here
# -- To ask about a specific function
?janitor::clean_names
?dplyr::group_byCongrats! You are all done! 🙌🏻
R coding tips and tricks
Useful keyboard shortcuts in RStudio
Description |
Windows & Linux |
Mac |
|---|---|---|
Insert code section |
Ctrl+Shift+R |
Shift+Command+R or Ctrl+Shift+R |
Insert code chunk (Quarto/Rmarkdown) |
Ctrl+Alt+I |
Command+Option+I |
Comment/uncomment line |
Ctrl+Shift+C |
Command+Shift+C |
Reindent lines |
Ctrl+I |
Command+I |
Insert 'assign' operator <- |
Alt+- |
Option+- |
Insert 'pipe' operator %>% or |> |
Ctrl+Shift+M |
Shift+Command+M |
Code completion (in source) |
Tab |
Tab |
File path completion (in console) |
"+Tab |
"+Tab |
Multple cursor selection |
Alt+click |
Alt+click |
Multple cursor selection (next) |
Alt+Shift+click |
Alt+Shift+click |
Switch cursor between source & console |
Ctrl+1 and Ctrl+2 |
Ctrl+1 and Ctrl+2 |
Run current line/selection |
Ctrl+Enter |
Command+Return |
Run current chunk |
Ctrl+Alt+C |
Command+Option+C |
Run entire file |
Ctrl+Shift+Enter |
Command+Shift+Return |
Show help for function at cursor |
F1 |
F1 |
Search within file |
Ctrl+F |
Command+F |
Search within project |
Ctrl+Shift+F |
Command+Shift+F |
Restart R session |
Ctrl+Shift+F10 |
Command+Shift+F10 |
Here you find the complete list of RStudio Keyboard Shortcuts